The WHO has declared that antimicrobial resistance (AMR) is one of the top 10 global public health threats facing humanity, and antimicrobial resistant infections may become the leading cause of death globally by 2050.
The Vivli AMR Register was successfully launched in June 2022 and is sharing surveillance data from GSK, Innoviva Specialty Therapeutics, Johnson & Johnson, Merck, Paratek, Pfizer, Shionogi, Venatorx and Venus Remedies, and welcomes data from other potential data contributors in pharmaceutical, diagnostic, biotech and generics companies who generate surveillance data.
The open sharing of these industry data through a single platform enables researchers to:
- Detect trends in multi-drug resistance over time
- Inform national and international policy, and antibiotic stewardship
- Allow modelling of future resistance trends
Note: all the datasets on the AMR Register are available as part of the challenge, with the exception of the Merck datasets. These datasets from Merck can still be requested via the AMR Register, but not as part of the data challenge.
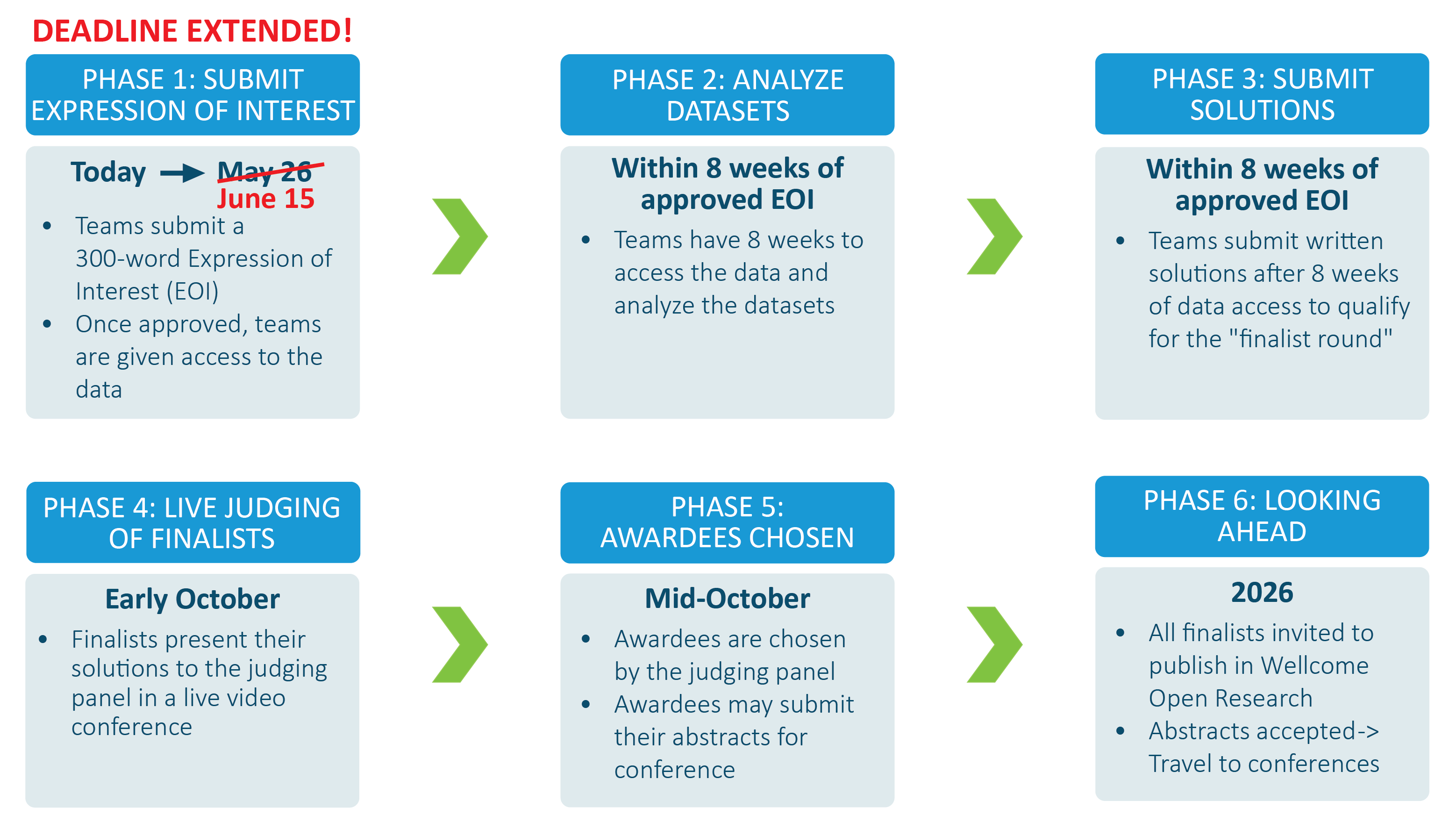
2025 Vivli AMR Surveillance Challenge, funded by Johnson & Johnson, Paratek, Pfizer, and an NIH award*
Our hope is that the research projects enabled by this platform will further advance understanding, inform decision-making and drive policy and behavioral changes in the medical community, by global organizations and wider society.
The key goal of the 2025 Vivli AMR Surveillance Data Challenge is to promote utilization of the Vivli AMR Register to more researchers and drive advances in the AMR field. We believe that important questions in surveillance would be answered, or tools developed through this Data Challenge.
This Challenge is in direct alignment with Wellcome** and Vivli’s open science goals in that through this project it will:
- Maximize the reach and discoverability of research
- Maximize the potential for research to be used and have real world impact
- Facilitate collaboration, engagement and dialogue around research
- Expand the user-base of the AMR surveillance data available on the AMR Register
To see the potential of the data, see the 2024 Data Challenge winners solutions and the 2023 Data Challenge winners solutions.
* Funded in part by NIH Award
Generalist Repository Ecosystem Initiative (GREI)
Other Transaction Agreement No.: 1OT2DB000003-01
**Wellcome funded the launch of the AMR Register and the 2023 Vivli AMR Surveillance Data Challenge.